使用R语言分析白榆生长数据
老王 / 2017-09-23
1. 数据初步整理
使用白榆生长数据。指标分别有:品系号、生长地点、环境盐碱度(低中高三种)、树高、胸径、东西冠幅、南北冠幅,为2017年测定。
在excel里整理数据,导出为csv格式,存在R工作目录下,进行操作。如果在windows下,使用read.csv 即可,但苹果中需要另外一种形式。
后来发现是自己忘了加引号了。。。
all.data = read.table("data for analysis/all.data.demo.txt", header = T, sep="\t", fileEncoding="UTF-16")
head(all.data)## X spe high dbh ew sn loca salt
## 1 1 58 5.04 6.2 2.12 2.90 河口 低
## 2 2 58 4.85 6.0 1.86 2.05 河口 低
## 3 3 58 4.83 6.3 2.18 2.42 河口 低
## 4 4 58 5.25 8.0 3.20 3.11 河口 低
## 5 5 58 5.35 8.0 2.98 2.92 河口 低
## 6 6 58 6.10 7.7 2.52 2.38 河口 低# 然后把第一列去掉
all.data = all.data[,-1]需要注意的是数据的格式,要以数据库能够认识的格式保存数据,即标题占一行,每一个个体指标都是独立,如上面所示。在excel里操作要注意千万不要合并单元格,最好把数据整理成数据透视表能够识别的格式。
对于excel中已经和并的数据,整理时有一个小技巧,假设列名是合并的:1、取消合并。2、选中列,F5定位。3、定位条件选空值。4、在第一个单元格里填写“=上一个单元格(地址)”。5、ctrl+enter。
数据涉及沾化、河口、垦利三地的1790棵树。包括变量 spe, high, dbh, ew, sn, loca, salt。
接下来分别整理并探索几个地方的数据的结构,按地方,按品系,按盐分梯度,不同角度都试试。
kenli = subset(all.data, loca == "垦利")
hekou = subset(all.data, loca == "河口")
zhanhua = subset(all.data, loca == "沾化")
# 把几个地方的数据分解出来
str(kenli)## 'data.frame': 622 obs. of 7 variables:
## $ spe : int 58 58 58 58 58 58 58 58 58 58 ...
## $ high: num 7.6 7.5 7.3 7.5 7.4 7.5 7.1 7.1 7 7 ...
## $ dbh : num 10.2 8.7 8.7 9.7 9.2 9.4 8.9 8.65 7.21 8.38 ...
## $ ew : num 3.98 3.19 3.29 3.8 3.5 3.47 3.87 4.13 3.12 4 ...
## $ sn : num 4.5 3.71 3.49 3.63 3.8 3.56 3.96 3.99 3.66 3.55 ...
## $ loca: Factor w/ 3 levels "垦利","河口",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ salt: Factor w/ 3 levels "中","低","高": 2 2 2 2 2 2 2 2 2 2 ...str(hekou)## 'data.frame': 1027 obs. of 7 variables:
## $ spe : int 58 58 58 58 58 58 58 58 58 58 ...
## $ high: num 5.04 4.85 4.83 5.25 5.35 6.1 5.7 5.03 4.85 4.95 ...
## $ dbh : num 6.2 6 6.3 8 8 7.7 8.3 7 5.4 5.4 ...
## $ ew : num 2.12 1.86 2.18 3.2 2.98 2.52 2.93 2.3 2.25 2.74 ...
## $ sn : num 2.9 2.05 2.42 3.11 2.92 2.38 3.18 2.17 1.28 2.03 ...
## $ loca: Factor w/ 3 levels "垦利","河口",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ salt: Factor w/ 3 levels "中","低","高": 2 2 2 2 2 2 2 2 1 1 ...str(zhanhua)## 'data.frame': 141 obs. of 7 variables:
## $ spe : int 58 58 58 58 58 58 58 58 58 58 ...
## $ high: num 4.5 4.7 4.8 4.8 4.8 3.9 4.1 4.05 4.1 4.1 ...
## $ dbh : num 6.4 7.18 5.35 7.54 5.93 4.35 5.55 4.43 3.68 4.2 ...
## $ ew : num 2.9 2.7 2.65 2.62 2.43 2.37 2.42 2.67 3.35 2.71 ...
## $ sn : num 3 3.4 3 3.19 2.93 2.55 2.77 2.35 3.3 3.23 ...
## $ loca: Factor w/ 3 levels "垦利","河口",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ salt: Factor w/ 3 levels "中","低","高": 3 3 3 3 3 3 3 3 3 3 ...3 排序(主成分分析)
3.1 第一次尝试
library(vegan)## Loading required package: permute## Loading required package: lattice## This is vegan 2.5-5library(ggplot2)## Registered S3 methods overwritten by 'ggplot2':
## method from
## [.quosures rlang
## c.quosures rlang
## print.quosures rlangplot(rda(kenli[,c(2,3,4,5)]))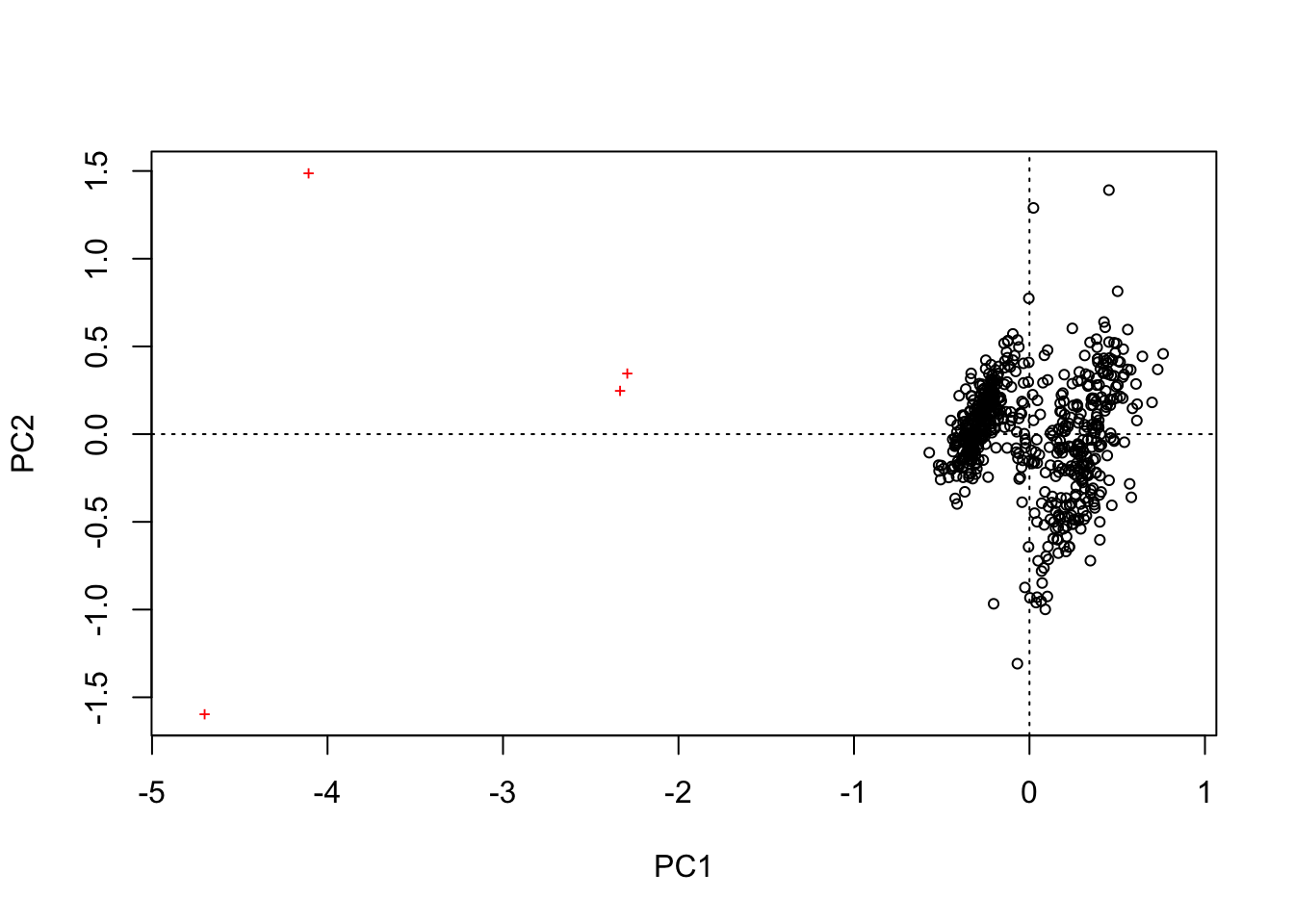
可以发现,点太多,区分不开。所以使用平均值进行分析。
这个过程可以在excel里完成,比较直观,也可以在R语言里完成。下面使用R语言。
3.2 把数据按品种算出平均值
library(dplyr) # 一个牛逼的包##
## Attaching package: 'dplyr'## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, unionhekou.spe = group_by(hekou, spe)
hekou.mean = summarise(hekou.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
head(hekou.mean)## # A tibble: 6 x 5
## spe high dbh ew sn
## <int> <dbl> <dbl> <dbl> <dbl>
## 1 5 5.33 5.92 2.16 2.12
## 2 7 5.25 6.19 2.21 2.14
## 3 20 5.27 6.01 2.19 2.19
## 4 30 5.37 6.54 2.44 2.30
## 5 34 5.16 5.88 2.17 2.03
## 6 41 5.23 5.94 2.09 2.09# 以河口为例,先分组,再按分组计算,和excel里的数据透视表有点像。
kenli.spe = group_by(kenli, spe)
kenli.mean = summarise(kenli.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
zhanhua.spe = group_by(zhanhua, spe)
zhanhua.mean = summarise(zhanhua.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)有了这些分组的平均值,可以再次尝试做主成分分析。
3.3 再次排序
library(tidyverse) #这个包是个组合,里面包括dplyr,ggplot2等一系列包。## ── Attaching packages ────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──## ✔ tibble 2.1.3 ✔ purrr 0.3.2
## ✔ tidyr 0.8.3 ✔ stringr 1.4.0
## ✔ readr 1.3.1 ✔ forcats 0.4.0## ── Conflicts ───────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()library(vegan)
hekou.mean = as.data.frame(hekou.mean)
hekou.mean = hekou.mean %>% remove_rownames %>% column_to_rownames(var="spe")
# %>% 是管道函数的符号,意思就是这么干了再这么干,比如上面就是先去掉行名,再把spe这一列变成行名。
# 如果直接做下面的函数,会有Warning message:
# Setting row names on a tibble is deprecated. 所以先把几个数据变成数据框。以垦利为例,原本差异如下:
kenli.mean## # A tibble: 12 x 5
## spe high dbh ew sn
## <int> <dbl> <dbl> <dbl> <dbl>
## 1 5 5.87 7.48 3.09 3.33
## 2 7 5.81 7.28 3.00 3.15
## 3 20 5.67 7.12 3.01 3.14
## 4 30 5.89 7.66 3.11 3.08
## 5 34 6.00 7.37 3.04 2.95
## 6 41 6.01 7.41 3.07 3.06
## 7 46 6.35 7.51 3.15 3.09
## 8 50 5.90 7.50 3.16 3.29
## 9 51 5.83 7.32 2.91 2.99
## 10 58 6.05 7.48 3.10 3.07
## 11 286 5.97 7.68 3.02 3.16
## 12 65225 6.15 7.42 3.06 2.96as.data.frame(kenli.mean)## spe high dbh ew sn
## 1 5 5.874000 7.484400 3.094400 3.327600
## 2 7 5.810000 7.280200 3.004600 3.150800
## 3 20 5.666667 7.123333 3.011000 3.144000
## 4 30 5.894444 7.659815 3.106111 3.076481
## 5 34 6.004167 7.369375 3.039375 2.947917
## 6 41 6.014118 7.411176 3.074314 3.057451
## 7 46 6.350000 7.513196 3.145652 3.092609
## 8 50 5.897959 7.499184 3.155306 3.294694
## 9 51 5.830508 7.315763 2.906271 2.990847
## 10 58 6.054000 7.484600 3.096000 3.069600
## 11 286 5.971667 7.677667 3.017333 3.161417
## 12 65225 6.153333 7.424222 3.060667 2.955778# 接着把垦利和沾化的数据也弄好。
kenli.mean = as.data.frame(kenli.mean)
kenli.mean = kenli.mean %>% remove_rownames %>% column_to_rownames(var="spe")
zhanhua.mean = as.data.frame(zhanhua.mean)
zhanhua.mean = zhanhua.mean %>% remove_rownames %>% column_to_rownames(var="spe")再做主成分分析。在vegan里,用rda函数。
hekou.rda = rda(hekou.mean)
str(hekou.rda)## List of 10
## $ colsum : Named num [1:4] 0.155 0.353 0.138 0.113
## ..- attr(*, "names")= chr [1:4] "high" "dbh" "ew" "sn"
## $ tot.chi : num 0.18
## $ Ybar : num [1:12, 1:4] 0.01233 -0.01169 -0.00681 0.02357 -0.04023 ...
## ..- attr(*, "scaled:center")= Named num [1:4] 5.29 6.12 2.21 2.15
## .. ..- attr(*, "names")= chr [1:4] "high" "dbh" "ew" "sn"
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:12] "5" "7" "20" "30" ...
## .. ..$ : chr [1:4] "high" "dbh" "ew" "sn"
## ..- attr(*, "METHOD")= chr "PCA"
## $ method : chr "rda"
## $ call : language rda(X = hekou.mean)
## $ pCCA : NULL
## $ CCA : NULL
## $ CA :List of 7
## ..$ eig : Named num [1:4] 0.166144 0.009104 0.004307 0.000869
## .. ..- attr(*, "names")= chr [1:4] "PC1" "PC2" "PC3" "PC4"
## ..$ poseig : NULL
## ..$ u : num [1:12, 1:4] 0.1387 -0.0307 0.0708 -0.3683 0.2186 ...
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:12] "5" "7" "20" "30" ...
## .. .. ..$ : chr [1:4] "PC1" "PC2" "PC3" "PC4"
## ..$ v : num [1:4, 1:4] -0.318 -0.861 -0.312 -0.245 -0.888 ...
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:4] "high" "dbh" "ew" "sn"
## .. .. ..$ : chr [1:4] "PC1" "PC2" "PC3" "PC4"
## ..$ rank : int 4
## ..$ tot.chi: num 0.18
## ..$ Xbar : num [1:12, 1:4] 0.01233 -0.01169 -0.00681 0.02357 -0.04023 ...
## .. ..- attr(*, "scaled:center")= Named num [1:4] 5.29 6.12 2.21 2.15
## .. .. ..- attr(*, "names")= chr [1:4] "high" "dbh" "ew" "sn"
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:12] "5" "7" "20" "30" ...
## .. .. ..$ : chr [1:4] "high" "dbh" "ew" "sn"
## .. ..- attr(*, "METHOD")= chr "PCA"
## $ inertia : chr "variance"
## $ regularization: chr "this is a vegan::rda result object"
## - attr(*, "class")= chr [1:2] "rda" "cca"plot(hekou.rda)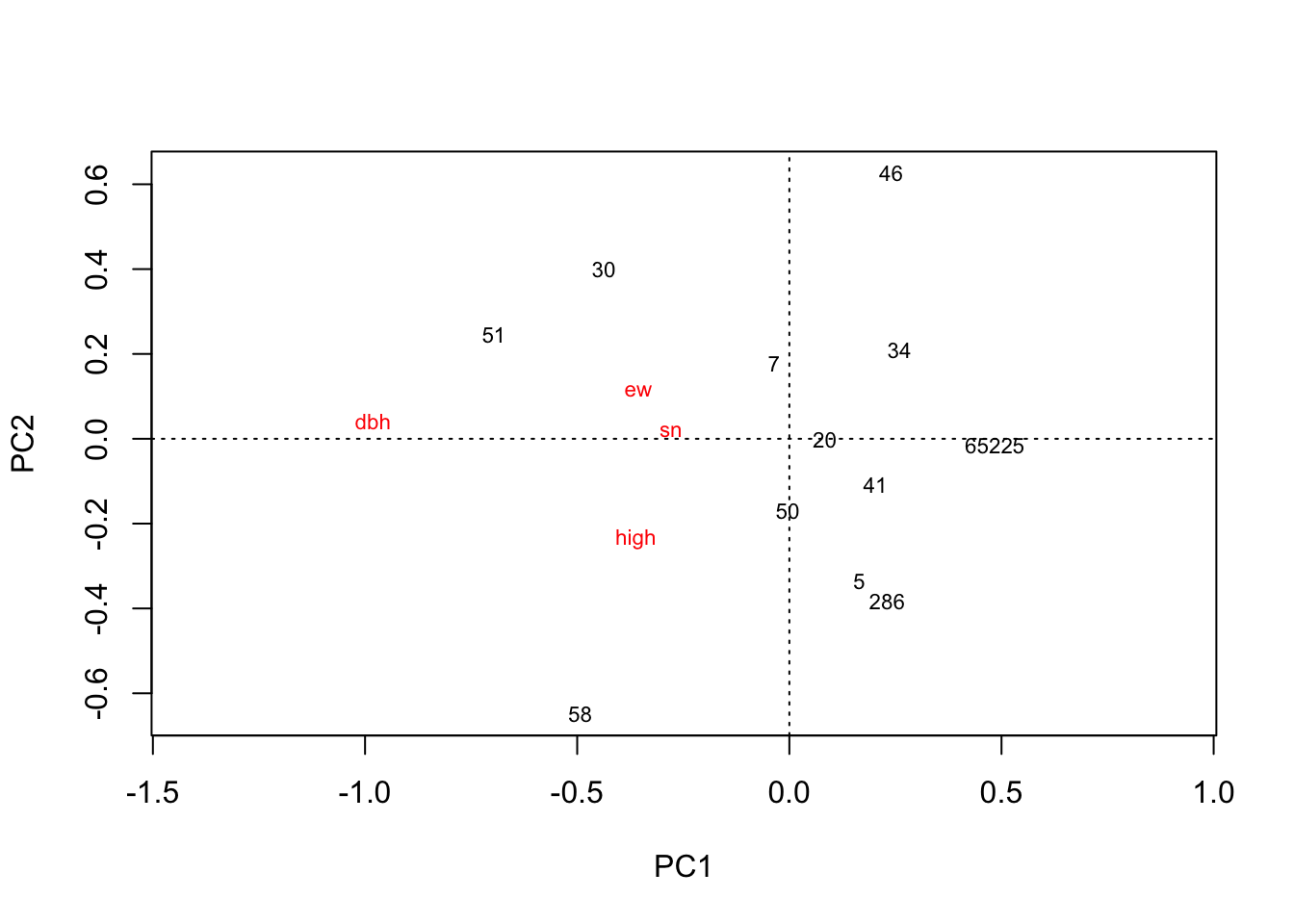
# 把需要作图的数据提取出来,取个好名字出现在坐标轴上
PC1 = hekou.rda$CA$u[,1]
PC2 = hekou.rda$CA$u[,2]
hekou.gg = as.data.frame(cbind(PC1, PC2))
ggplot(hekou.gg, aes(PC1, PC2, label = row.names(hekou.gg))) +
geom_point() +
geom_text( vjust = "inward", hjust = "inward")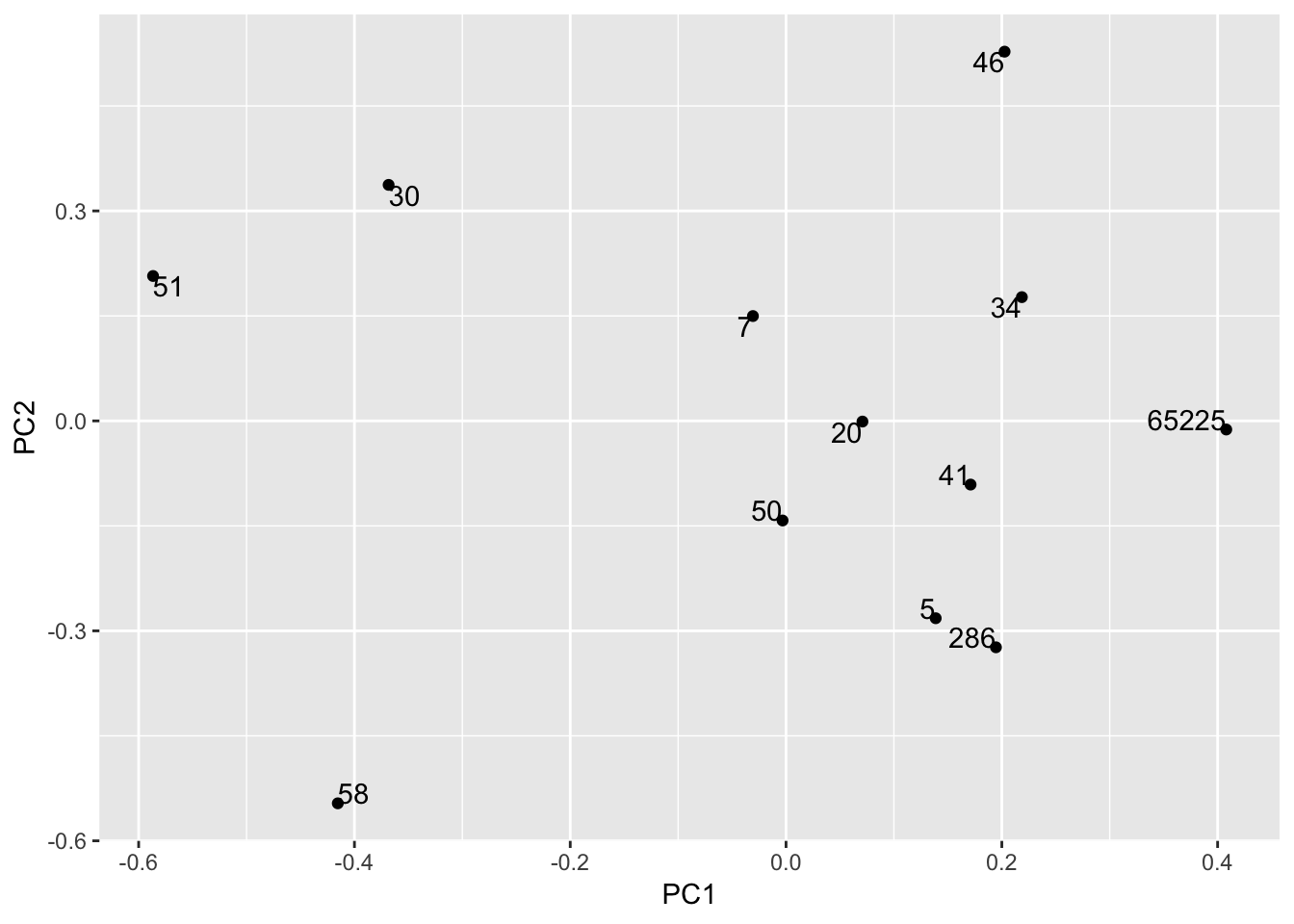
# 这个好看点,但不如vegan自带的作图容易说明问题。所以不用这个,仅作方法保存一下。
# 注意ggplot的用法,若要不同颜色,则需要把颜色对应变量转化为character或factor,默认为数值型,为渐变颜色。接下来,从不同角度作图,开始。
library(dplyr)
kenli.low = filter(all.data, loca == "垦利", salt == "低")
kenli.low.spe = group_by(kenli.low, spe)
kenli.low.mean = summarise(kenli.low.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
kenli.med = filter(all.data, loca == "垦利", salt == "中")
kenli.med.spe = group_by(kenli.med, spe)
kenli.med.mean = summarise(kenli.med.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
kenli.high = filter(all.data, loca == "垦利", salt == "高")
kenli.high.spe = group_by(kenli.high, spe)
kenli.high.mean = summarise(kenli.high.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
hekou.low = filter(all.data, loca == "河口", salt == "低")
hekou.low.spe = group_by(hekou.low, spe)
hekou.low.mean = summarise(hekou.low.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
hekou.med = filter(all.data, loca == "河口", salt == "中")
hekou.med.spe = group_by(hekou.med, spe)
hekou.med.mean = summarise(hekou.med.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
hekou.high = filter(all.data, loca == "河口", salt == "高")
hekou.high.spe = group_by(hekou.high, spe)
hekou.high.mean = summarise(hekou.high.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
zhanhua.low = filter(all.data, loca == "沾化", salt == "低")
zhanhua.low.spe = group_by(zhanhua.low, spe)
zhanhua.low.mean = summarise(zhanhua.low.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
zhanhua.med = filter(all.data, loca == "沾化", salt == "中")
zhanhua.med.spe = group_by(zhanhua.med, spe)
zhanhua.med.mean = summarise(zhanhua.med.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
zhanhua.high = filter(all.data, loca == "沾化", salt == "高")
zhanhua.high.spe = group_by(zhanhua.high, spe)
zhanhua.high.mean = summarise(zhanhua.high.spe,
high = mean(high),
dbh = mean(dbh),
ew = mean(ew),
sn = mean(sn)
)
kenli.low.mean = as.data.frame(kenli.low.mean)
kenli.low.mean = kenli.low.mean %>% remove_rownames %>% column_to_rownames(var="spe")
kenli.med.mean = as.data.frame(kenli.med.mean)
kenli.med.mean = kenli.med.mean %>% remove_rownames %>% column_to_rownames(var="spe")
kenli.high.mean = as.data.frame(kenli.high.mean)
kenli.high.mean = kenli.high.mean %>% remove_rownames %>% column_to_rownames(var="spe")
hekou.low.mean = as.data.frame(hekou.low.mean)
hekou.low.mean = hekou.low.mean %>% remove_rownames %>% column_to_rownames(var="spe")
hekou.med.mean = as.data.frame(hekou.med.mean)
hekou.med.mean = hekou.med.mean %>% remove_rownames %>% column_to_rownames(var="spe")
hekou.high.mean = as.data.frame(hekou.high.mean)
hekou.high.mean = hekou.high.mean %>% remove_rownames %>% column_to_rownames(var="spe")
zhanhua.low.mean = as.data.frame(zhanhua.low.mean)
zhanhua.low.mean = zhanhua.low.mean %>% remove_rownames %>% column_to_rownames(var="spe")
zhanhua.med.mean = as.data.frame(zhanhua.med.mean)
zhanhua.med.mean = zhanhua.med.mean %>% remove_rownames %>% column_to_rownames(var="spe")
zhanhua.high.mean = as.data.frame(zhanhua.high.mean)
zhanhua.high.mean = zhanhua.high.mean %>% remove_rownames %>% column_to_rownames(var="spe")作图了。
library(vegan)
plot(rda(kenli.low.mean))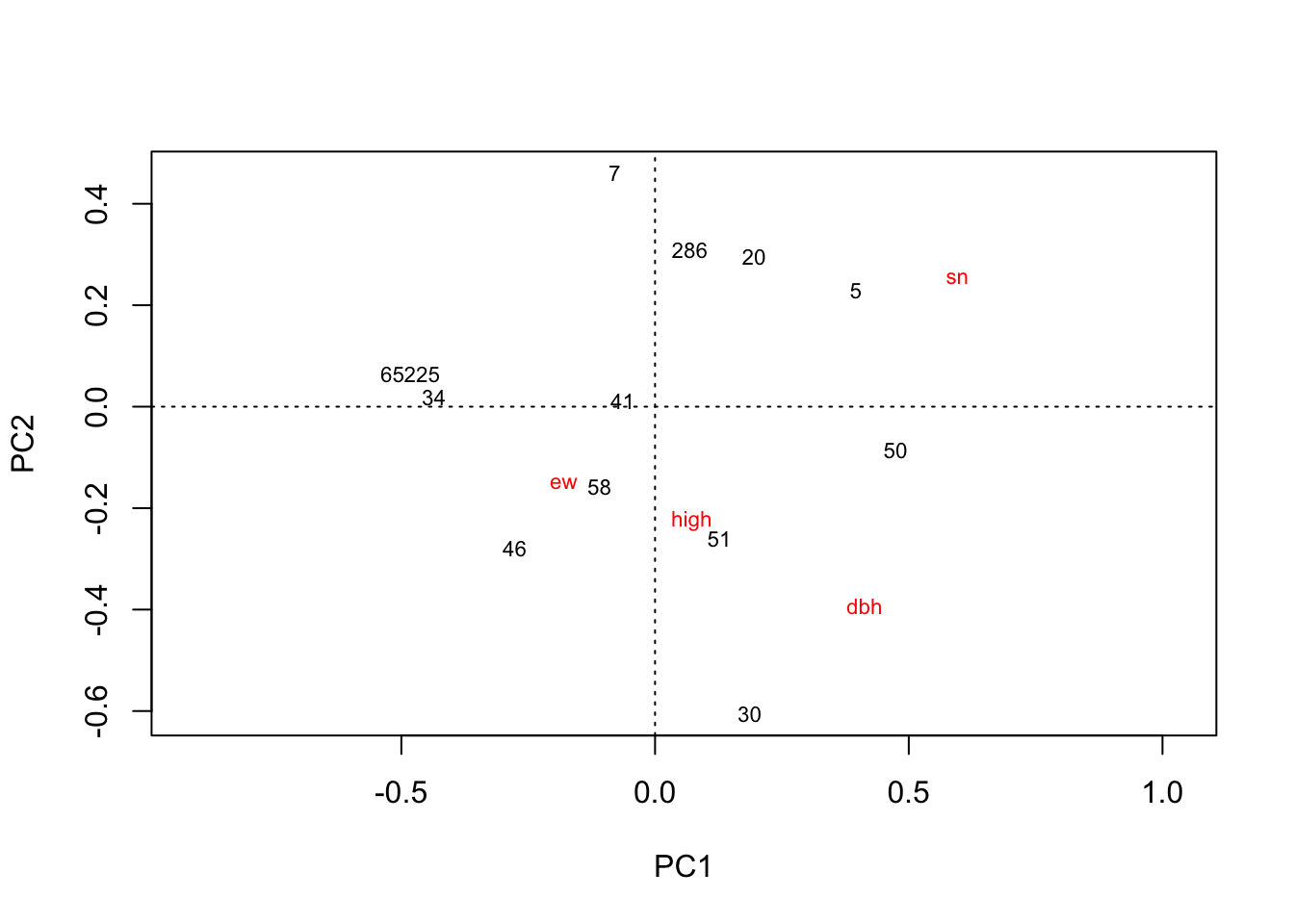
# 垦利低盐分
plot(rda(kenli.med.mean))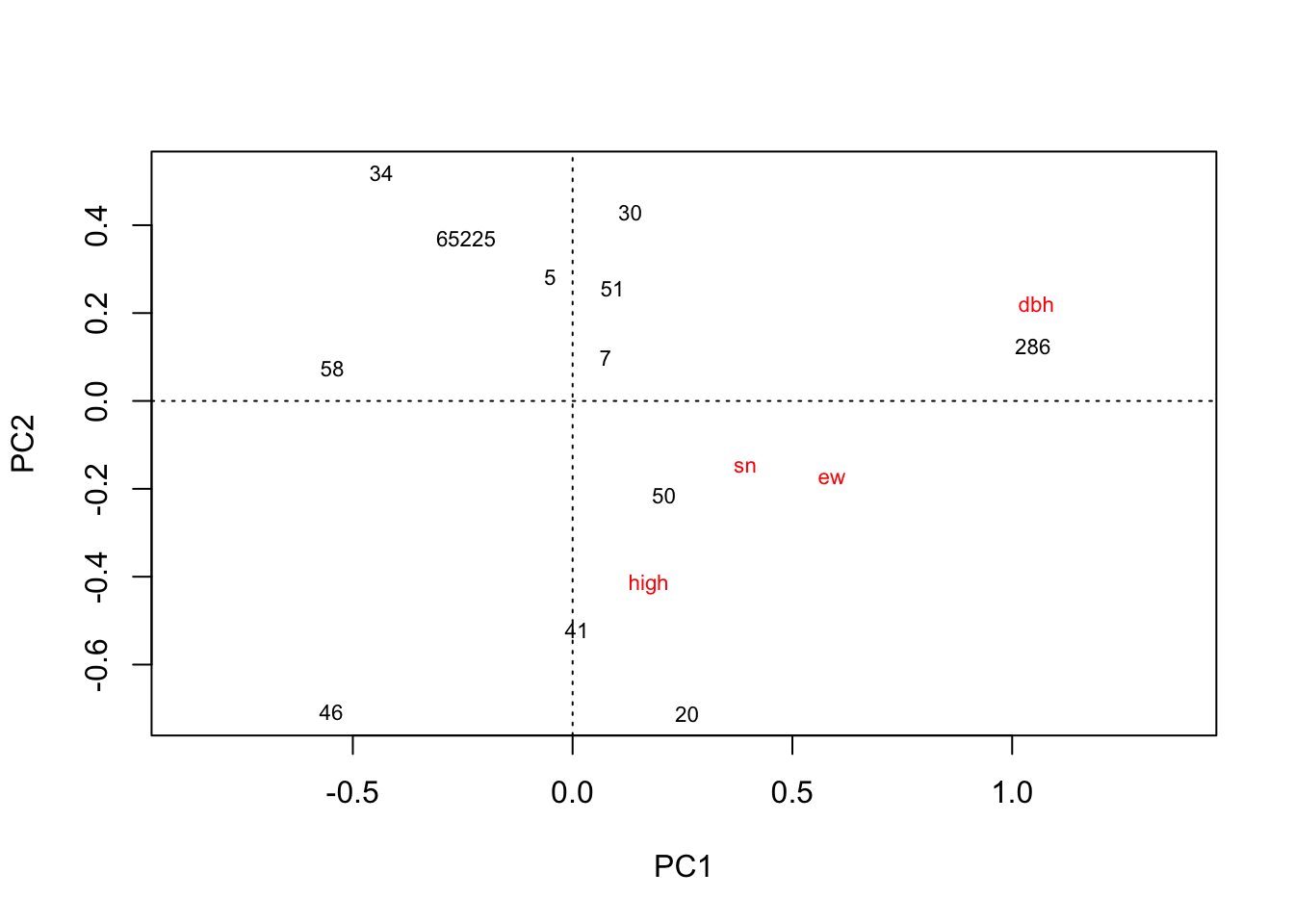
# 垦利中盐分
plot(rda(kenli.high.mean))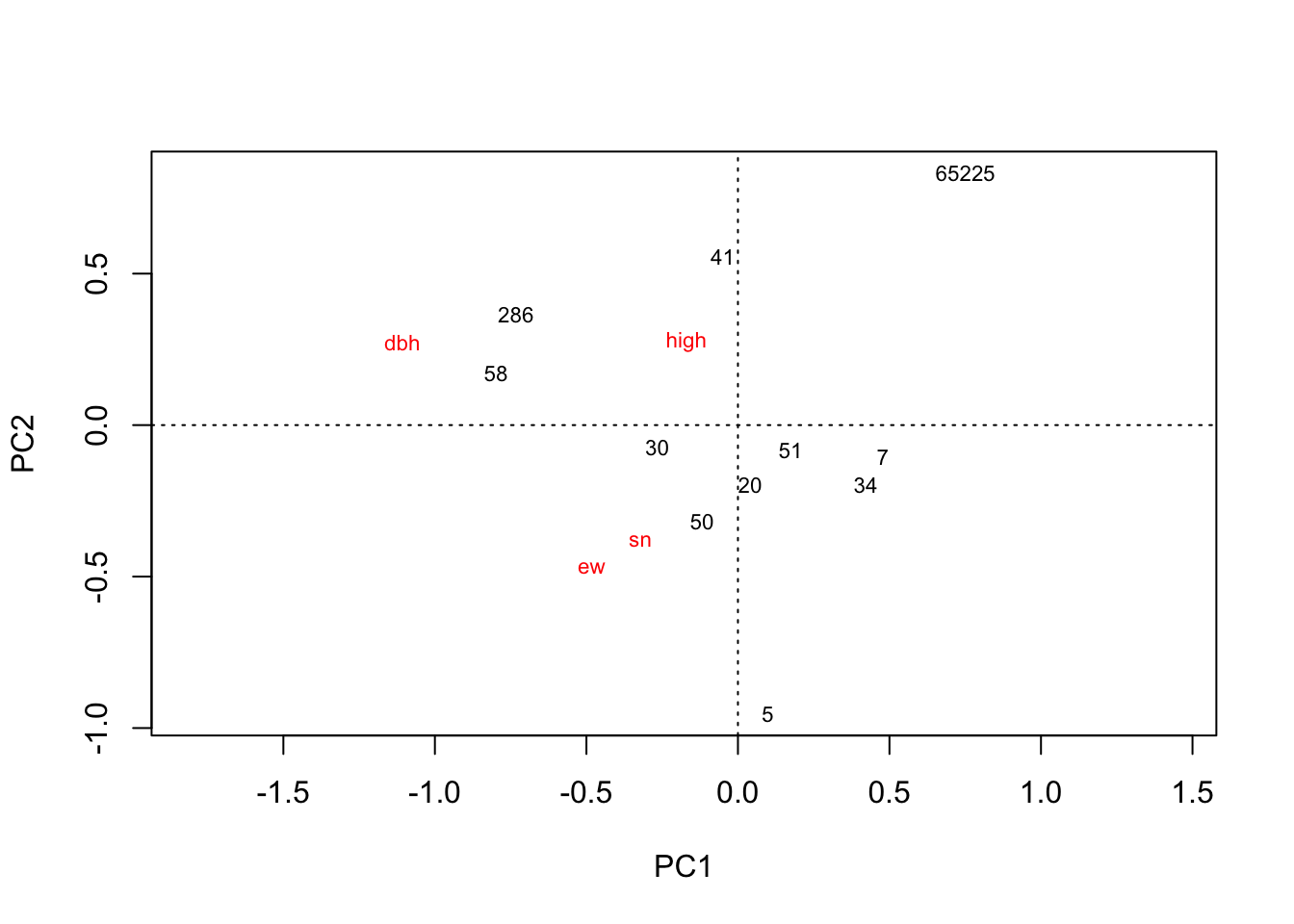
# 垦利高盐分
plot(rda(hekou.low.mean))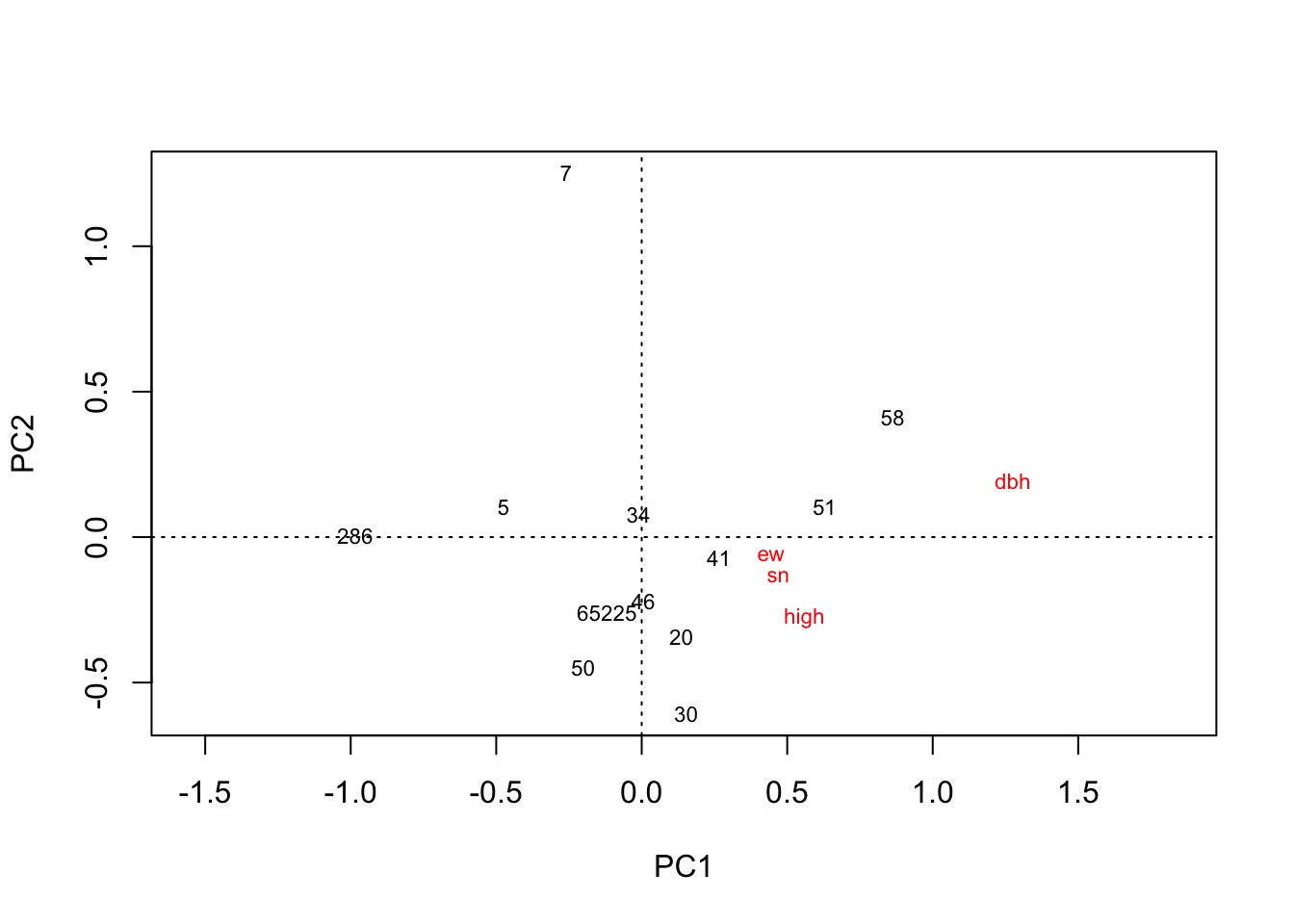
# 河口低盐分
plot(rda(hekou.med.mean))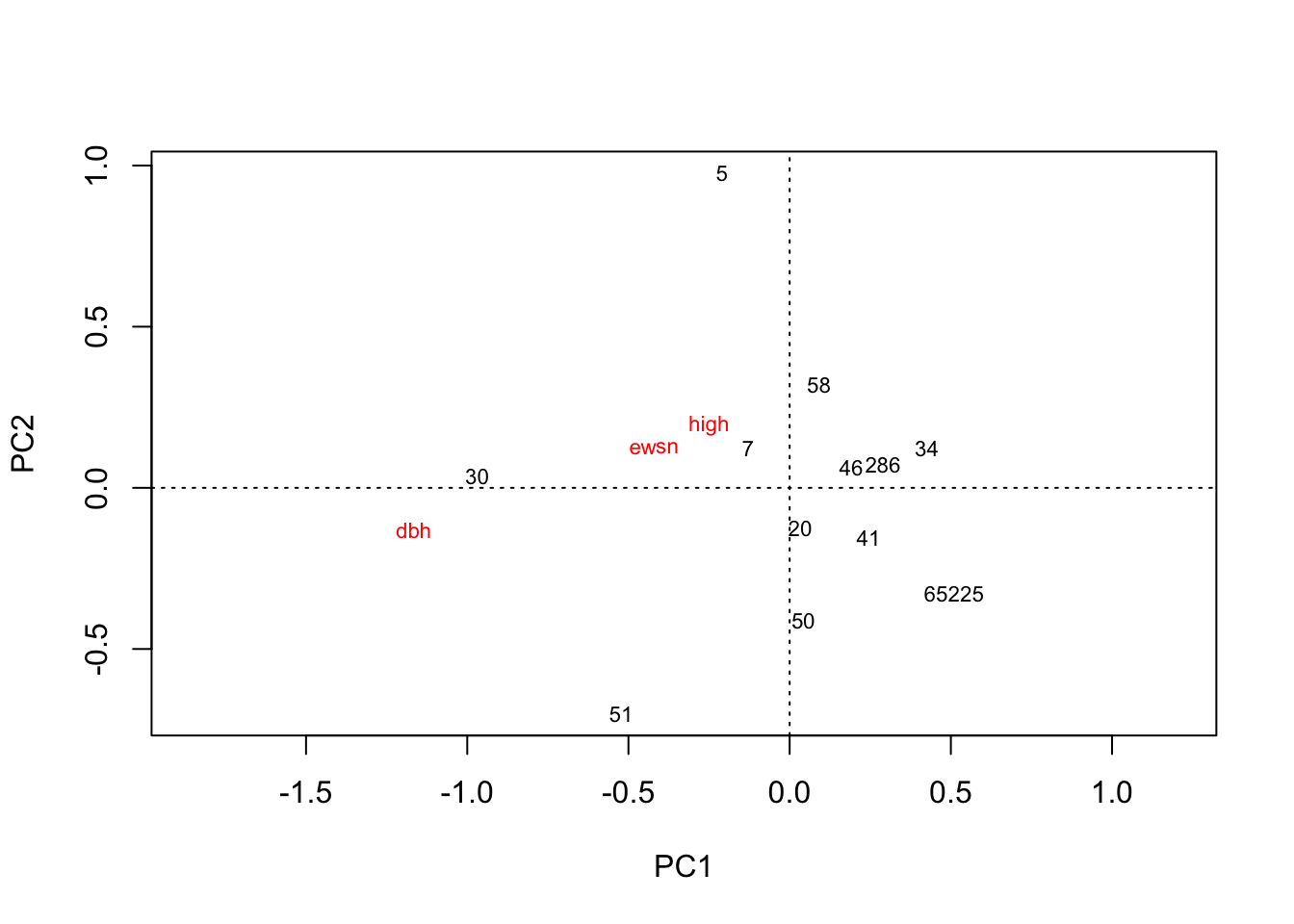
# 河口中盐分
plot(rda(hekou.high.mean))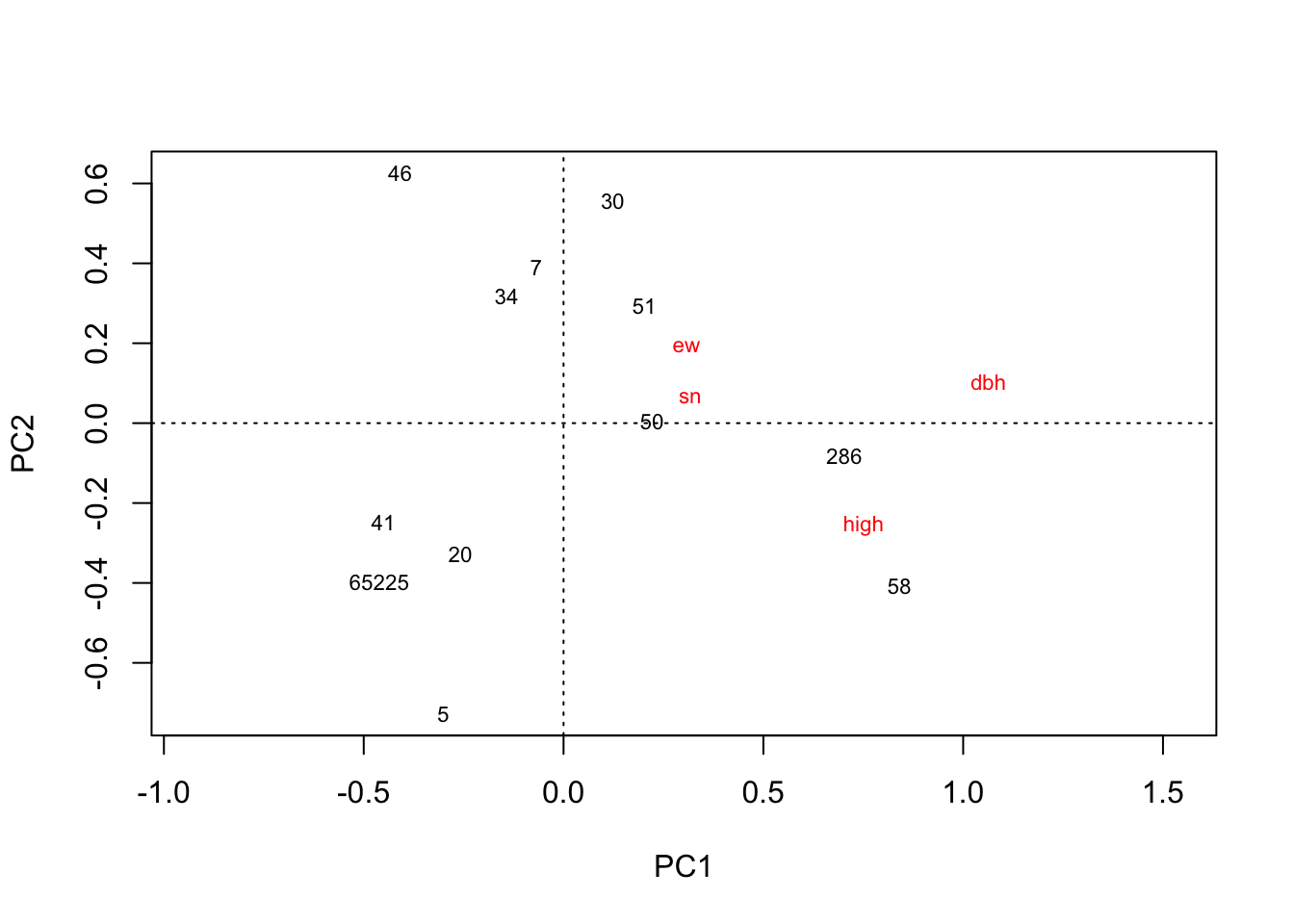
# 河口高盐分
plot(rda(zhanhua.high.mean))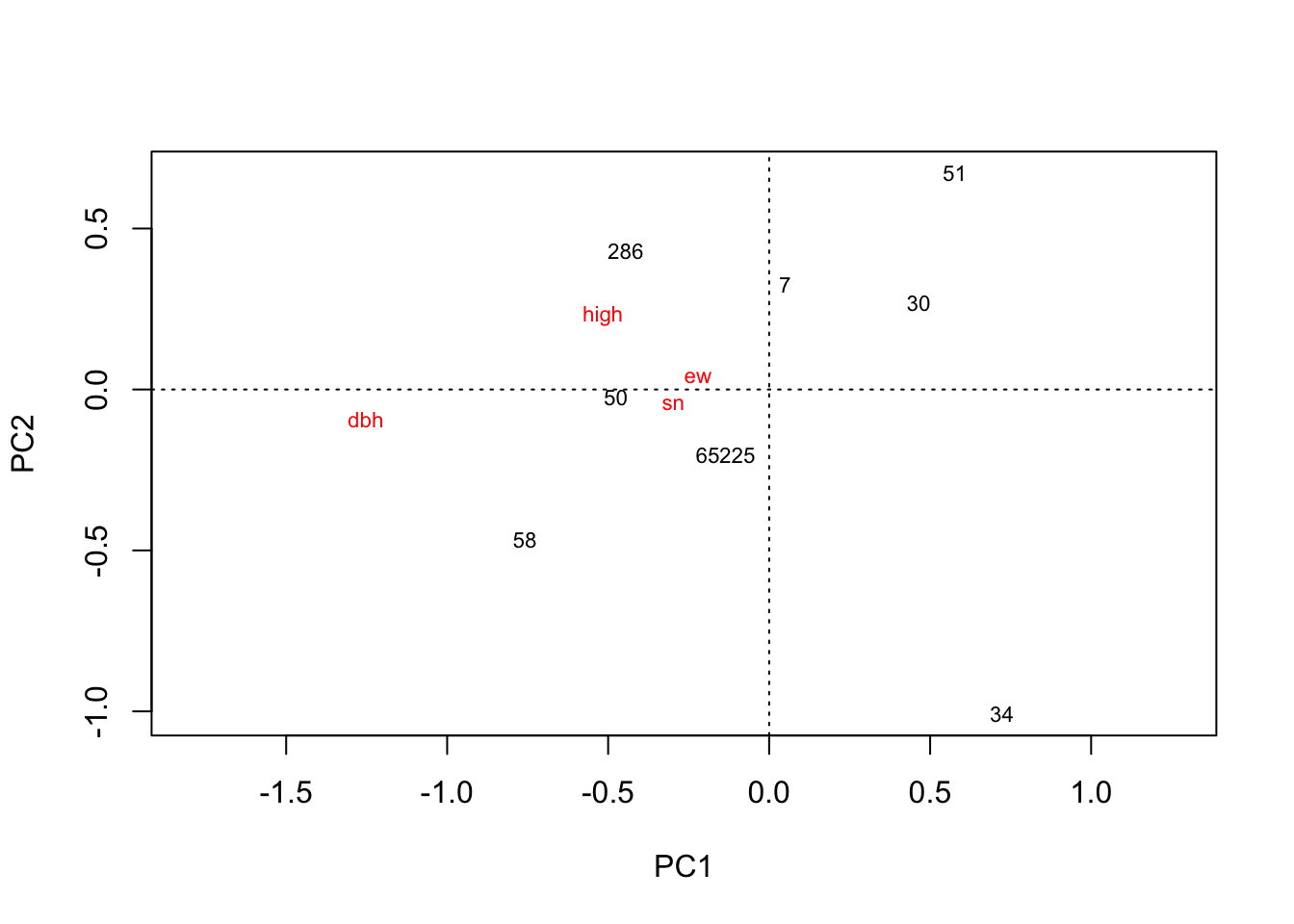
# 沾化高盐分4. 聚类分析
4.1 聚类分析方法的选择
聚类分析,要先算出样本间距离来,这里直接用平均值。如何体现重复数据需要进一步研究,存疑。
用的函数hclust,方法如下:
1, 数据标准化:decostand(data, “normalize”)
2, 计算欧式距离:vegdist(data, “euc”),或者dist()
3, 不同聚类方法
3.1 Hierarchical Clustering Based on Links Single Linkage Agglomerative Clustering: hclust(distance, method = “single”) Complet Linkage Agglomerative Clustering: hclust(distance, method = “complete”)
3.2 Average Agglomerative Clustering UPGMA best-known method: hclust(distance, method = “average”) 还有其他几种average clustering: UPGMC“centroid”, WPGMA“mcquitty”, WPGMC“median”
3.3 Ward’s Minimus Variance Clustering Based on the linear model criterion of least squares. method = “ward”
- 4,解释聚类结果
Cophenetic Correlation 表型相关,the distance where the two objects become
members of the same group.
聚类的case是每一行的名称,如果是直接在excel里读取的话,存成csv格式,用read.csv(“testdata.csv”, row.names=1)读取,里面数据均参与计算相似系数矩阵。
目前觉得single方法是最合适的,是最近相邻法聚类,能够较好的把不同类别的品种分为多类。
4.2 聚类作图
dis.kenli.high = vegdist(decostand(kenli.high.mean, "normalize"), "euc")
dis.kenli.med = vegdist(decostand(kenli.med.mean, "normalize"), "euc")
dis.kenli.low = vegdist(decostand(kenli.low.mean, "normalize"), "euc")
kenli.high.single = hclust(dis.kenli.high, method = "single")
kenli.med.single = hclust(dis.kenli.med, method = "single")
kenli.low.single = hclust(dis.kenli.low, method = "single")
plot(kenli.high.single)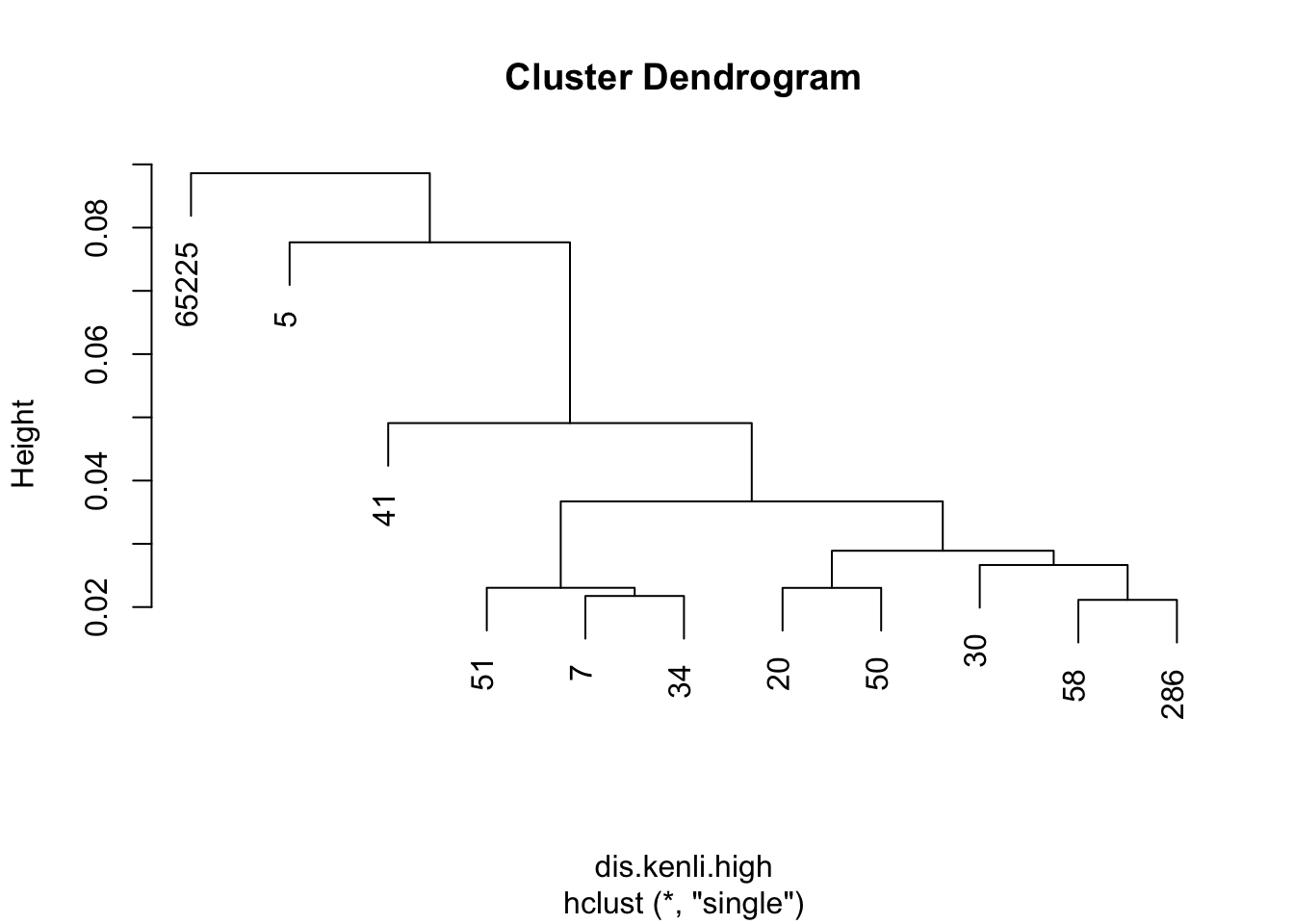
plot(kenli.med.single)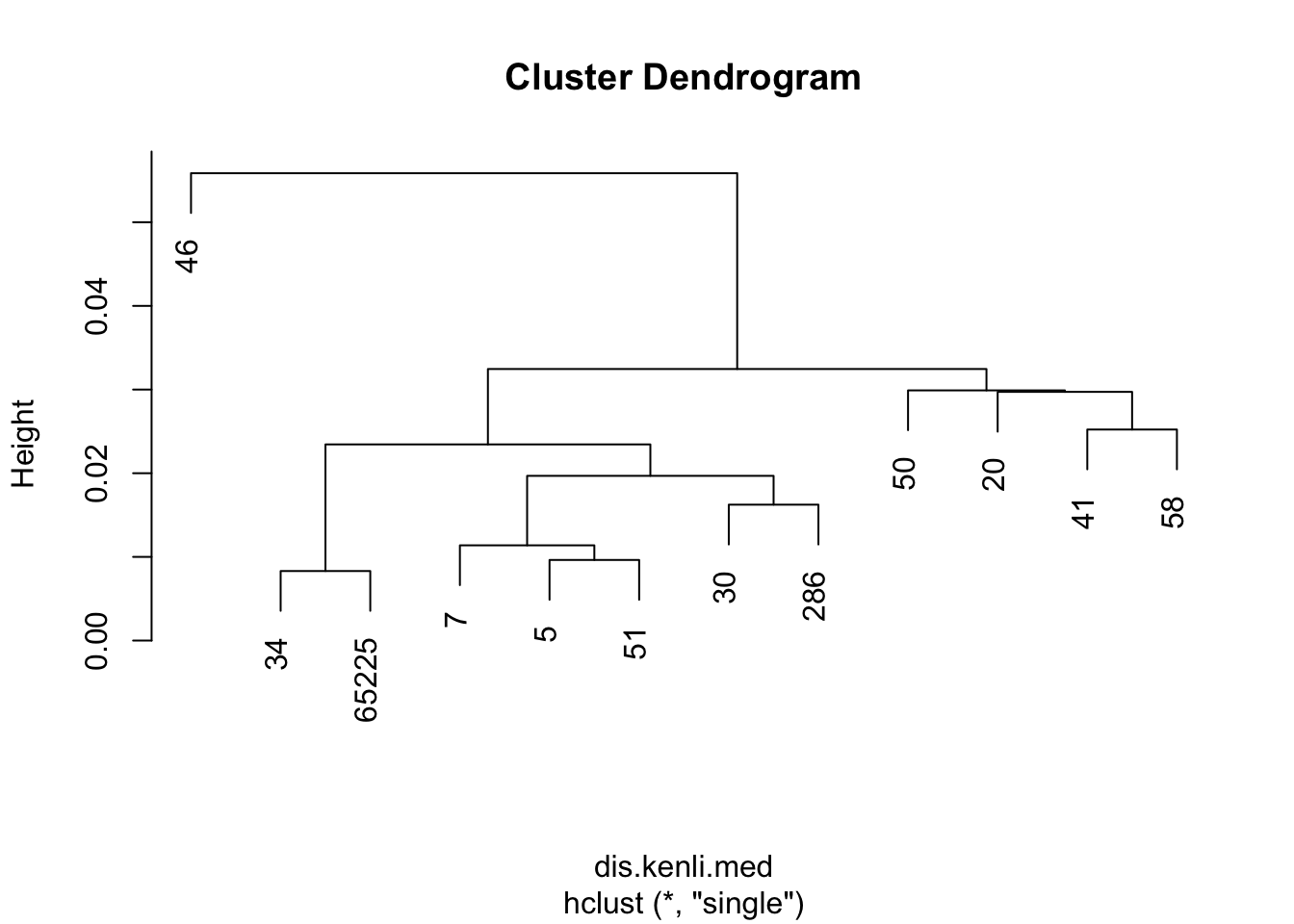
plot(kenli.low.single)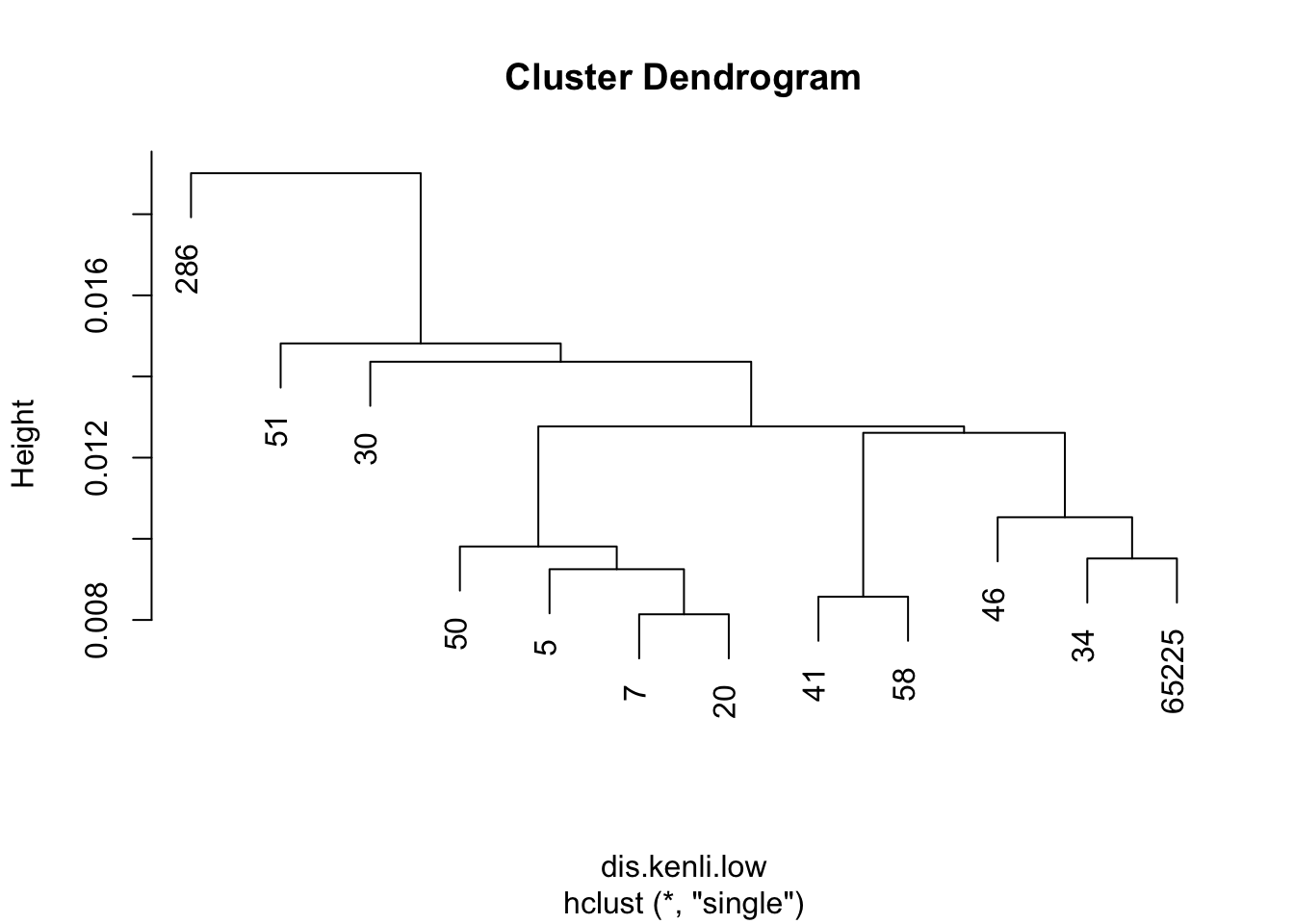
5 小结
- 树高、胸径、冠幅有时候并不完全一致,使用PCA能从整体的角度找到优良品种;聚类可以直观把这些品种分成不同的团伙。
- 数据整理过程中,对r语言不熟悉的话,不如用数据透视表快。
- 以上分析为了试试用blogdown写博客并用hugo挂网上,检验图能出现,格式正确。
- 真是太方便了,R分析,网站表达,直接打通了。